Non-Small Cell Lung Cancer Drug Resistance
We are a research group, led by Prof. Hong Yan, working on the analysis of EGFR mutations and drug resistance related to non-small cell lung cancer (NSCLC). We are interested in building computational models, designing effective algorithms, and developing machine learning techniques to solve key open problems, including predicting 3D structures of EGFR mutants, understanding the mechanisms of drug resistance, and proposing methods to combat drug resistance and personalized treatment of NSCLC patients.
We have collaborations with medical doctors, biochemists, biologists and mathematicians at hospitals and universities. We have received the following joint research grants:
Health and Medical Research Fund (HMRF): Computational Platform for Modelling, Analysis and Prediction of Anti-EGFR Drug Resistance for Lung Cancer.
Project investigators: Hong Yan (EE, CityU), Guangyu Zhu (Chemistry, CityU), Maria Wong (Pathology, Queen Mary Hospital and HKU Faculty of Medicine), Victor Lee (Clinical Oncology, Queen Mary Hospital and HKU Faculty of Medicine)
Click here to see our project information on HMRF website.
Click here to see our project report on HMRF website.
Click here to see all known EGFR mutations discovered at Queen Mary Hospital and those reported in literature.
Click here to see our publications from the HMRF project.
Click here to see comments on our work from other medical doctors and researchers.
Collaborative Research Fund (CRF): Efficient Algorithms and Hardware Accelerators for Tensor Decomposition and Their Applications to Multidimensional Data Analysis.
Project investigators: Hong Yan (EE, CityU), Raymond Chan (Math, CUHK/CityU), Ray Cheung (EE, CityU), Victor Lee (Clinical Oncology, Queen Mary Hospital and HKU Faculty of Medicine), Michael NG (Math, HKBU), Liqun Qi (Applied Math, PolyU)
Click here to see fund information on RGC website (Project C1007-15G).
Click here to see project abstract on RGC website (Project C1007-15G).
IEEE Award: Professor Hong Yan received the 2016 Norbert Wiener Award from IEEE Systems, Man and Cybernetics Society for contributions to image and biomolecular pattern recognition techniques.
Click here to see the award certificate.
Click here to see CityU news report.
Lung cancer is the leading cause of cancer deaths worldwide. Non-small-cell lung cancer (NSCLC) constitutes about 85% of all lung cancer cases. Mutation of the epidermal growth factor receptor (EGFR) is a common cause of NSCLC and its incidence can reach 60% in East Asian population. The "driver" mutations increase the kinase activity of EGFR, leading to EGFR overexpression and uncontrolled lung cell division and eventually lung cancer.
Tyrosine-kinase inhibitors (TKI), such as gefitinib, erlotinib and osimertinib, against EGFR-mutated NSCLC, have been developed and investigated extensively in the past two decades. These inhibitors bind to the EGFR kinase domain, which blocks the subsequent signal transduction leading to inhibition of tumor proliferation. Tumors with activating EGFR mutations, especially deletions in exon 19 and L858R in exon 21, are particularly responsive to these TKIs leading to a longer overall survival (OS) rate of pateints compared to systemic chemotherapy. Despite initial promising and dramatic response to these TKIs, drug resistance will ultimately develop with almost every NSCLC patient within one to two years.
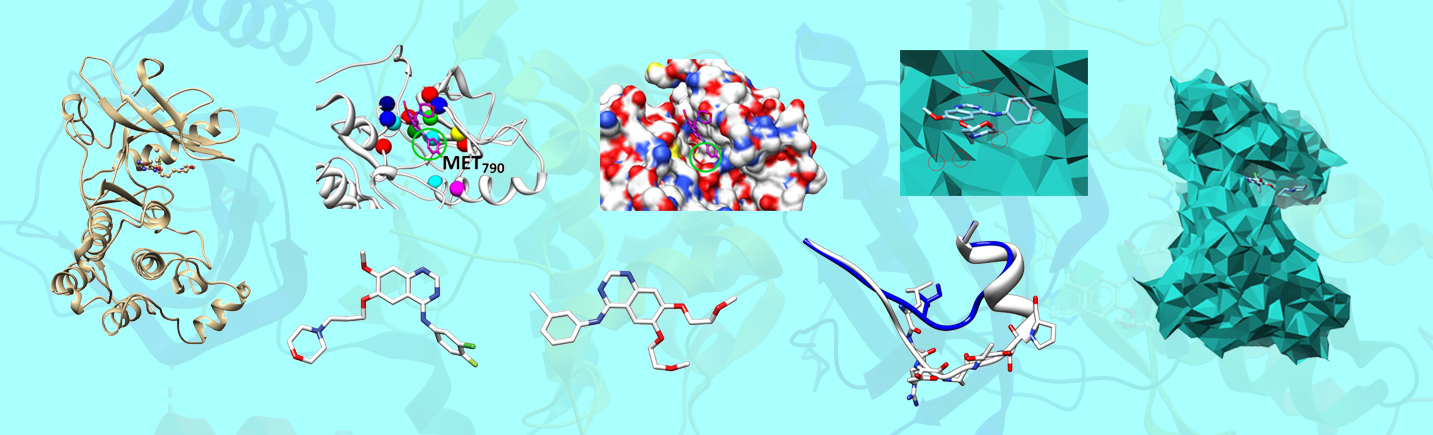
Structural information is available for just a few EGFR mutants. We created an EGFR Mutant Structural Database for all known EGFR mutants, which can be used by other researchers to study NSCLC further and by medical doctors as reference for NSCLC treatment.
For a certain TKI, patients with some EGFR mutants may response to this TKI while patients with other mutants may not. For example, patients with EGFR mutations of deletions in exon 19 or L858R in exon 21 will response well to first-generation TKIs gefitinib and erlotinib, but patients with the mutation T790M will resistant to these two TKIs. On the other hand, for patients with a specific EGFR mutation, some TKIs may work well while others may not. For example, patients with the mutation T790M will have a good response to the third-generation TKI osimertinib.
Our group has developed several innovative methods to investigate the drug resistance problem. We have studied local surface geometric properties of EGFR mutants based on 3D alpha shape models, characteristics of EGFR-drug binding patterns based on eigen-binding site models, and the formation of EGFR related homo-dimers and hetero-dimers. Our research can provide useful computational tools for medical doctors to plan personalized treatment of NSCLC patients.